Dystrophin
| DMD | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | DMD, BMD, CMD3B, DXS142, DXS164, DXS206, DXS230, DXS239, DXS268, DXS269, DXS270, DXS272, MRX85, dystrophin | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 300377 MGI: 94909 HomoloGene: 20856 GeneCards: DMD | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||

Dystrophin is a rod-shaped cytoplasmic protein, and a vital part of a protein complex that connects the cytoskeleton of a muscle fiber to the surrounding extracellular matrix through the cell membrane. This complex is variously known as the costamere or the dystrophin-associated protein complex (DAPC). Many muscle proteins, such as α-dystrobrevin, syncoilin, synemin, sarcoglycan, dystroglycan, and sarcospan, colocalize with dystrophin at the costamere. It has a molecular weight of 427 kDa [5][6]
Dystrophin is coded for by the DMD gene – the largest known human gene, covering 2.4 megabases (0.08% of the human genome) at locus Xp21. The primary transcript in muscle measures about 2,100 kilobases and takes 16 hours to transcribe;[7] the mature mRNA measures 14.0 kilobases.[8] The 79-exon muscle transcript[9] codes for a protein of 3685 amino acid residues.[10]
Spontaneous or inherited mutations in the dystrophin gene can cause different forms of muscular dystrophy, a disease characterized by progressive muscular wasting. The most common of these disorders caused by genetic defects in dystrophin is Duchenne muscular dystrophy.
Function
Dystrophin is a protein located between the sarcolemma and the outermost layer of myofilaments in the muscle fiber (myofiber). It is a cohesive protein, linking actin filaments to other support proteins that reside on the inside surface of each muscle fiber's plasma membrane (sarcolemma). These support proteins on the inside surface of the sarcolemma in turn links to two other consecutive proteins for a total of three linking proteins. The final linking protein is attached to the fibrous endomysium of the entire muscle fiber. Dystrophin supports muscle fiber strength, and the absence of dystrophin reduces muscle stiffness, increases sarcolemmal deformability, and compromises the mechanical stability of costameres and their connections to nearby myofibrils. This has been shown in recent studies where biomechanical properties of the sarcolemma and its links through costameres to the contractile apparatus were measured,[11] and helps to prevent muscle fiber injury. Movement of thin filaments (actin) creates a pulling force on the extracellular connective tissue that eventually becomes the tendon of the muscle. The dystrophin associated protein complex also helps scaffold various signalling and channel proteins, implicating the DAPC in regulation of signalling processes.[12]
Pathology
Dystrophin deficiency has been definitively established as one of the root causes of the general class of myopathies collectively referred to as muscular dystrophy. The deletions of one or several exons of the dystrophin DMD gene cause Duchenne and Becker muscular dystrophies.[13] The large cytosolic protein was first identified in 1987 by Louis M. Kunkel,[14] after concurrent works by Kunkel and Robert G. Worton to characterize the mutated gene that causes Duchenne muscular dystrophy (DMD).[15][16] At least nine disease-causing mutations in this gene have been discovered.[17]
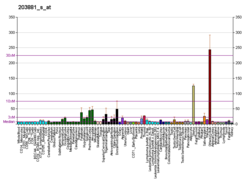
Normal skeletal muscle tissue contains only small amounts of dystrophin (about 0.002% of total muscle protein),[14] but its absence (or abnormal expression) leads to the development of a severe and currently incurable constellation of symptoms most readily characterized by several aberrant intracellular signaling pathways that ultimately yield pronounced myofiber necrosis as well as progressive muscle weakness and fatigability. Most DMD patients become wheelchair-dependent early in life, and the gradual development of cardiac hypertrophy—a result of severe myocardial fibrosis—typically results in premature death in the first two or three decades of life. Variants (mutations) in the DMD gene that lead to the production of too little or a defective, internally shortened but partially functional dystrophin protein, result in a display of a much milder dystrophic phenotype in affected patients, resulting in the disease known as Becker's muscular dystrophy (BMD). In some cases, the patient's phenotype is such that experts may decide differently on whether a patient should be diagnosed with DMD or BMD. The theory currently most commonly used to predict whether a variant will result in a DMD or BMD phenotype, is the reading frame rule.[18]
Though its role in airway smooth muscle is not well established, recent research indicates that dystrophin along with other subunits of dystrophin glycoprotein complex is associated with phenotype maturation.[19]
Research
A number of models are used to facilitate research on DMD gene defects. These include the mdx mouse, GRMD (golden retriever muscular dystrophy) dog, and HFMD (hypertrophic feline muscular dystrophy) cat.[20]
The mdx mouse contains a nonsense mutation in exon 23, leading to a shortened dystrophin protein.[21] Levels of dystrophin in this model is not zero: a variety of mutation alleles exist with measurable levels certain of dystrophin isoforms.[20] Muscle degeneration pathology is most easily visible in the diaphragm.[22] Generally, clinically relevant pathology is observed with older mdx mice.[22]
The GRMD dog is one of several existing dystrophin-deficient dogs identified where substantial characterization has been performed.[23] Clinically relevant pathology can be observed at 8 weeks after birth, with continued gradual deterioration of muscle function.[24] Muscle histology is most analogous to clinical presentation of DMD in humans with necrosis, fibrosis and regeneration.[25]
The HFMD cat has a deletion in the promoter region of the DMD gene.[26] Muscle histology shows necrosis but no fibrosis.[27] Extensive hypertrophy has been observed which is thought to be responsible for shorter lifespans.[28][27] Due to the hypertrophy, this model may have limited uses for DMD studies.
Therapeutic microdystrophin
- Delandistrogene moxeparvovec - systemic gene transfer with rAAVrh74.MHCK7.micro-dystrophin.[29][30][31]
Interactions
Dystrophin has been shown to interact with:
Neanderthal admixture
A variant of the DMD gene, which is on the X chromosome, named B006, appears to be an introgression from a Neanderthal-modern human mating.[37]
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000198947 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000045103 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Lederfein D, Levy Z, Augier N, Mornet D, Morris G, Fuchs O, et al. (June 1992). "A 71-kilodalton protein is a major product of the Duchenne muscular dystrophy gene in brain and other nonmuscle tissues". Proceedings of the National Academy of Sciences of the United States of America. 89 (12): 5346–50. Bibcode:1992PNAS...89.5346L. doi:10.1073/pnas.89.12.5346. PMC 49288. PMID 1319059.
- ^ "DMD - Dystrophin - Homo sapiens (Human) - DMD gene & protein". www.uniprot.org. Retrieved 1 December 2021.
- ^ Tennyson CN, Klamut HJ, Worton RG (February 1995). "The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced". Nature Genetics. 9 (2): 184–90. doi:10.1038/ng0295-184. PMID 7719347. S2CID 7858296.
- ^ NCBI Sequence Viewer v2.0
- ^ Strachan T and Read AP, 1999. Human molecular genetics, BIOS Scientific, New York, USA
- ^ "dystrophin isoform Dp427c [Homo sapiens] - Protein - NCBI". www.ncbi.nlm.nih.gov.
- ^ García-Pelagio KP, Bloch RJ, Ortega A, González-Serratos H (March 2011). "Biomechanics of the sarcolemma and costameres in single skeletal muscle fibers from normal and dystrophin-null mice". Journal of Muscle Research and Cell Motility. 31 (5–6): 323–36. doi:10.1007/s10974-011-9238-9. PMC 4326082. PMID 21312057.
- ^ Constantin B (February 2014). "Dystrophin complex functions as a scaffold for signalling proteins". Biochimica et Biophysica Acta (BBA) - Biomembranes. 1838 (2): 635–42. doi:10.1016/j.bbamem.2013.08.023. PMID 24021238.
- ^ Le Rumeur E (July 2015). "Dystrophin and the two related genetic diseases, Duchenne and Becker muscular dystrophies". Bosnian Journal of Basic Medical Sciences. 15 (3): 14–20. doi:10.17305/bjbms.2015.636. PMC 4594321. PMID 26295289.
- ^ a b Hoffman EP, Brown RH, Kunkel LM (December 1987). "Dystrophin: the protein product of the Duchenne muscular dystrophy locus". Cell. 51 (6): 919–28. doi:10.1016/0092-8674(87)90579-4. PMID 3319190. S2CID 33548364.
- ^ Monaco AP, Neve RL, Colletti-Feener C, Bertelson CJ, Kurnit DM, Kunkel LM (1986). "Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene". Nature. 323 (6089): 646–50. Bibcode:1986Natur.323..646M. doi:10.1038/323646a0. PMID 3773991. S2CID 4317085.
- ^ Burghes AH, Logan C, Hu X, Belfall B, Worton RG, Ray PN (1987). "A cDNA clone from the Duchenne/Becker muscular dystrophy gene". Nature. 328 (6129): 434–7. Bibcode:1987Natur.328..434B. doi:10.1038/328434a0. PMID 3614347. S2CID 4364629.
- ^ Šimčíková D, Heneberg P (December 2019). "Refinement of evolutionary medicine predictions based on clinical evidence for the manifestations of Mendelian diseases". Scientific Reports. 9 (1): 18577. Bibcode:2019NatSR...918577S. doi:10.1038/s41598-019-54976-4. PMC 6901466. PMID 31819097.
- ^ Aartsma-Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJ, Den Dunnen JT (August 2006). "Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule". Muscle & Nerve. 34 (2): 135–44. doi:10.1002/mus.20586. PMID 16770791. S2CID 42303520.
- ^ Sharma P, Tran T, Stelmack GL, McNeill K, Gosens R, Mutawe MM, Unruh H, Gerthoffer WT, Halayko AJ (January 2008). "Expression of the dystrophin-glycoprotein complex is a marker for human airway smooth muscle phenotype maturation". American Journal of Physiology. Lung Cellular and Molecular Physiology. 294 (1): L57–68. doi:10.1152/ajplung.00378.2007. PMID 17993586.
- ^ a b Blake DJ, Weir A, Newey SE, Davies KE (April 2002). "Function and genetics of dystrophin and dystrophin-related proteins in muscle". Physiological Reviews. 82 (2): 291–329. doi:10.1152/physrev.00028.2001. PMID 11917091.
- ^ Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, Barnard PJ (June 1989). "The molecular basis of muscular dystrophy in the mdx mouse: a point mutation". Science. 244 (4912): 1578–80. Bibcode:1989Sci...244.1578S. doi:10.1126/science.2662404. PMID 2662404.
- ^ a b Stedman HH, Sweeney HL, Shrager JB, Maguire HC, Panettieri RA, Petrof B, et al. (August 1991). "The mdx mouse diaphragm reproduces the degenerative changes of Duchenne muscular dystrophy". Nature. 352 (6335): 536–9. Bibcode:1991Natur.352..536S. doi:10.1038/352536a0. PMID 1865908. S2CID 4302451.
- ^ "Duchenne Muscular Dystrophy and Becker Muscular Dystrophy: Diagnostic Principles". Duchenne Muscular Dystrophy. CRC Press. 2006-02-27. pp. 105–118. doi:10.3109/9780849374456-7. ISBN 978-0-429-16351-7.
- ^ Valentine BA, Cooper BJ, de Lahunta A, O'Quinn R, Blue JT (December 1988). "Canine X-linked muscular dystrophy. An animal model of Duchenne muscular dystrophy: clinical studies". Journal of the Neurological Sciences. 88 (1–3): 69–81. doi:10.1016/0022-510X(88)90206-7. PMID 3225630. S2CID 6902011.
- ^ Valentine BA, Cooper BJ, Cummings JF, de Lahunta A (June 1990). "Canine X-linked muscular dystrophy: morphologic lesions". Journal of the Neurological Sciences. 97 (1): 1–23. doi:10.1016/0022-510x(90)90095-5. PMID 2370557. S2CID 31250421.
- ^ Winand NJ, Edwards M, Pradhan D, Berian CA, Cooper BJ (September 1994). "Deletion of the dystrophin muscle promoter in feline muscular dystrophy". Neuromuscular Disorders. 4 (5–6): 433–45. doi:10.1016/0960-8966(94)90082-5. PMID 7881288. S2CID 38556669.
- ^ a b Carpenter JL, Hoffman EP, Romanul FC, Kunkel LM, Rosales RK, Ma NS, et al. (November 1989). "Feline muscular dystrophy with dystrophin deficiency". The American Journal of Pathology. 135 (5): 909–19. PMC 1880103. PMID 2683799.
- ^ Gaschen FP, Hoffman EP, Gorospe JR, Uhl EW, Senior DF, Cardinet GH, Pearce LK (July 1992). "Dystrophin deficiency causes lethal muscle hypertrophy in cats". Journal of the Neurological Sciences. 110 (1–2): 149–59. doi:10.1016/0022-510x(92)90022-d. PMID 1506854. S2CID 21156994.
- ^ "Chugai In-Licenses Gene Therapy Delandistrogene Moxeparvovec (SRP-9001) for Duchenne Muscular Dystrophy | News". 16 December 2021.
- ^ Mendell JR, Sahenk Z, Lehman K, Nease C, Lowes LP, Miller NF, et al. (September 2020). "Assessment of Systemic Delivery of rAAVrh74.MHCK7.micro-dystrophin in Children With Duchenne Muscular Dystrophy: A Nonrandomized Controlled Trial". JAMA Neurology. 77 (9): 1122–1131. doi:10.1001/jamaneurol.2020.1484. PMC 7296461. PMID 32539076.
- ^ "Delandistrogene moxeparvovec - Roche/Sarepta Therapeutics". AdisInsight. Springer Nature Switzerland AG.
- ^ Sadoulet-Puccio HM, Rajala M, Kunkel LM (November 1997). "Dystrobrevin and dystrophin: an interaction through coiled-coil motifs". Proceedings of the National Academy of Sciences of the United States of America. 94 (23): 12413–8. Bibcode:1997PNAS...9412413S. doi:10.1073/pnas.94.23.12413. PMC 24974. PMID 9356463.
- ^ Ahn AH, Freener CA, Gussoni E, Yoshida M, Ozawa E, Kunkel LM (February 1996). "The three human syntrophin genes are expressed in diverse tissues, have distinct chromosomal locations, and each bind to dystrophin and its relatives". The Journal of Biological Chemistry. 271 (5): 2724–30. doi:10.1074/jbc.271.5.2724. PMID 8576247.
- ^ Yang B, Jung D, Rafael JA, Chamberlain JS, Campbell KP (March 1995). "Identification of alpha-syntrophin binding to syntrophin triplet, dystrophin, and utrophin". The Journal of Biological Chemistry. 270 (10): 4975–8. doi:10.1074/jbc.270.10.4975. PMID 7890602.
- ^ Gee SH, Madhavan R, Levinson SR, Caldwell JH, Sealock R, Froehner SC (January 1998). "Interaction of muscle and brain sodium channels with multiple members of the syntrophin family of dystrophin-associated proteins". The Journal of Neuroscience. 18 (1): 128–37. doi:10.1523/jneurosci.18-01-00128.1998. PMC 6793384. PMID 9412493.
- ^ Ahn AH, Kunkel LM (February 1995). "Syntrophin binds to an alternatively spliced exon of dystrophin". The Journal of Cell Biology. 128 (3): 363–71. doi:10.1083/jcb.128.3.363. PMC 2120343. PMID 7844150.
- ^ Khan R (January 25, 2011). "Neandertal admixture, revisiting results after shaken priors". Discover Magazine. Archived from the original on January 27, 2013. Retrieved March 27, 2013.
Further reading
- Roberts RG, Gardner RJ, Bobrow M (1994). "Searching for the 1 in 2,400,000: a review of dystrophin gene point mutations". Human Mutation. 4 (1): 1–11. doi:10.1002/humu.1380040102. PMID 7951253. S2CID 24596547.
- Tinsley JM, Blake DJ, Zuellig RA, Davies KE (August 1994). "Increasing complexity of the dystrophin-associated protein complex". Proceedings of the National Academy of Sciences of the United States of America. 91 (18): 8307–13. Bibcode:1994PNAS...91.8307T. doi:10.1073/pnas.91.18.8307. PMC 44595. PMID 8078878.
- Blake DJ, Weir A, Newey SE, Davies KE (April 2002). "Function and genetics of dystrophin and dystrophin-related proteins in muscle". Physiological Reviews. 82 (2): 291–329. doi:10.1152/physrev.00028.2001. PMID 11917091.
- Röper K, Gregory SL, Brown NH (November 2002). "The 'spectraplakins': cytoskeletal giants with characteristics of both spectrin and plakin families". Journal of Cell Science. 115 (Pt 22): 4215–25. doi:10.1242/jcs.00157. hdl:2440/41876. PMID 12376554.
- Muntoni F, Torelli S, Ferlini A (December 2003). "Dystrophin and mutations: one gene, several proteins, multiple phenotypes". The Lancet. Neurology. 2 (12): 731–40. doi:10.1016/S1474-4422(03)00585-4. PMID 14636778. S2CID 34532766.
- Haenggi T, Fritschy JM (July 2006). "Role of dystrophin and utrophin for assembly and function of the dystrophin glycoprotein complex in non-muscle tissue" (PDF). Cellular and Molecular Life Sciences. 63 (14): 1614–31. doi:10.1007/s00018-005-5461-0. PMID 16710609. S2CID 8580596.
External links

- GeneReviews/NCBI/NIH/UW entry on Dystrophinopathies
- Dystrophin at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- LOVD mutation database: DMD, DMD (whole exon changes)
- v
- t
- e
-
 1dxx: N-TERMINAL ACTIN-BINDING DOMAIN OF HUMAN DYSTROPHIN
1dxx: N-TERMINAL ACTIN-BINDING DOMAIN OF HUMAN DYSTROPHIN -
 1eg3: STRUCTURE OF A DYSTROPHIN WW DOMAIN FRAGMENT IN COMPLEX WITH A BETA-DYSTROGLYCAN PEPTIDE
1eg3: STRUCTURE OF A DYSTROPHIN WW DOMAIN FRAGMENT IN COMPLEX WITH A BETA-DYSTROGLYCAN PEPTIDE -
 1eg4: STRUCTURE OF A DYSTROPHIN WW DOMAIN FRAGMENT IN COMPLEX WITH A BETA-DYSTROGLYCAN PEPTIDE
1eg4: STRUCTURE OF A DYSTROPHIN WW DOMAIN FRAGMENT IN COMPLEX WITH A BETA-DYSTROGLYCAN PEPTIDE