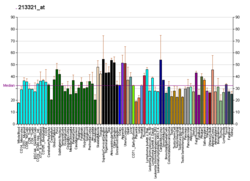
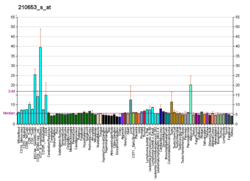
BCKDHB
| BCKDHB | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | BCKDHB, E1B, dJ279A18.1, BCKDE1B, BCKDH E1-beta, branched chain keto acid dehydrogenase E1, beta polypeptide, branched chain keto acid dehydrogenase E1 subunit beta | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 248611; MGI: 88137; HomoloGene: 39; GeneCards: BCKDHB; OMA:BCKDHB - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
2-Oxoisovalerate dehydrogenase subunit beta, mitochondrial is an enzyme that in humans is encoded by the BCKDHB gene.[5]
Function
Branched-chain keto acid dehydrogenase is a multienzyme complex associated with the inner membrane of mitochondria, and functions in the catabolism of branched-chain amino acids. The complex consists of multiple copies of 3 components: branched-chain alpha-keto acid decarboxylase (E1), lipoamide acyltransferase (E2), and lipoamide dehydrogenase (E3). This gene encodes the E1 beta subunit, and mutations therein have been associated with maple syrup urine disease (MSUD), type 1B. Alternative splicing at this locus results in transcript variants with different 3' noncoding regions, but encoding the same isoform.[5]
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000083123 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000032263 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b "Entrez Gene: BCKDHB branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease)".
Further reading
- Popov KM, Zhao Y, Shimomura Y, et al. (1992). "Branched-chain alpha-ketoacid dehydrogenase kinase. Molecular cloning, expression, and sequence similarity with histidine protein kinases". J. Biol. Chem. 267 (19): 13127–30. doi:10.1016/S0021-9258(18)42179-5. PMID 1377677.
- Mitsubuchi H, Nobukuni Y, Endo F, Matsuda I (1991). "Structural organization and chromosomal localization of the gene for the E1 beta subunit of human branched chain alpha-keto acid dehydrogenase". J. Biol. Chem. 266 (22): 14686–91. doi:10.1016/S0021-9258(18)98740-5. PMID 1860867.
- Zneimer SM, Lau KS, Eddy RL, et al. (1991). "Regional assignment of two genes of the human branched-chain alpha-keto acid dehydrogenase complex: the E1 beta gene (BCKDHB) to chromosome 6p21-22 and the E2 gene (DBT) to chromosome 1p31". Genomics. 10 (3): 740–7. doi:10.1016/0888-7543(91)90458-Q. PMID 1889817.
- Nobukuni Y, Mitsubuchi H, Akaboshi I, et al. (1991). "Maple syrup urine disease. Complete defect of the E1 beta subunit of the branched chain alpha-ketoacid dehydrogenase complex due to a deletion of an 11-bp repeat sequence which encodes a mitochondrial targeting leader peptide in a family with the disease". J. Clin. Invest. 87 (5): 1862–6. doi:10.1172/JCI115209. PMC 295312. PMID 2022752.
- Chuang JL, Cox RP, Chuang DT (1990). "Molecular cloning of the mature E1b-beta subunit of human branched-chain alpha-keto acid dehydrogenase complex". FEBS Lett. 262 (2): 305–9. Bibcode:1990FEBSL.262..305C. doi:10.1016/0014-5793(90)80215-5. PMID 2335211. S2CID 84069883.
- Nobukuni Y, Mitsubuchi H, Endo F, et al. (1990). "Maple syrup urine disease. Complete primary structure of the E1 beta subunit of human branched chain alpha-ketoacid dehydrogenase complex deduced from the nucleotide sequence and a gene analysis of patients with this disease". J. Clin. Invest. 86 (1): 242–7. doi:10.1172/JCI114690. PMC 296713. PMID 2365818.
- Wynn RM, Kochi H, Cox RP, Chuang DT (1994). "Differential processing of human and rat E1 alpha precursors of the branched-chain alpha-keto acid dehydrogenase complex caused by an N-terminal proline in the rat sequence". Biochim. Biophys. Acta. 1201 (1): 125–8. doi:10.1016/0304-4165(94)90161-9. PMID 7918575.
- Nobukuni Y, Mitsubuchi H, Hayashida Y, et al. (1993). "Heterogeneity of mutations in maple syrup urine disease (MSUD): screening and identification of affected E1 alpha and E1 beta subunits of the branched-chain alpha-keto-acid dehydrogenase multienzyme complex". Biochim. Biophys. Acta. 1225 (1): 64–70. doi:10.1016/0925-4439(93)90123-i. PMID 8161368.
- Chuang JL, Cox RP, Chuang DT (1996). "Maple syrup urine disease: the E1beta gene of human branched-chain alpha-ketoacid dehydrogenase complex has 11 rather than 10 exons, and the 3' UTR in one of the two E1beta mRNAs arises from intronic sequences". Am. J. Hum. Genet. 58 (6): 1373–7. PMC 1915070. PMID 8651316.
- Edelmann L, Wasserstein MP, Kornreich R, et al. (2001). "Maple syrup urine disease: identification and carrier-frequency determination of a novel founder mutation in the Ashkenazi Jewish population". Am. J. Hum. Genet. 69 (4): 863–8. doi:10.1086/323677. PMC 1226071. PMID 11509994.
- Chang CF, Chou HT, Chuang JL, et al. (2002). "Solution structure and dynamics of the lipoic acid-bearing domain of human mitochondrial branched-chain alpha-keto acid dehydrogenase complex". J. Biol. Chem. 277 (18): 15865–73. doi:10.1074/jbc.M110952200. PMID 11839747.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. USA. 99 (26): 16899–903. Bibcode:2002PNAS...9916899M. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Wynn RM, Machius M, Chuang JL, et al. (2003). "Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase: refined phosphorylation loop structure in the active site". J. Biol. Chem. 278 (44): 43402–10. doi:10.1074/jbc.M306204200. PMID 12902323.
- Li J, Wynn RM, Machius M, et al. (2004). "Cross-talk between thiamin diphosphate binding and phosphorylation loop conformation in human branched-chain alpha-keto acid decarboxylase/dehydrogenase". J. Biol. Chem. 279 (31): 32968–78. doi:10.1074/jbc.M403611200. PMID 15166214.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Machius M, Wynn RM, Chuang JL, et al. (2006). "A versatile conformational switch regulates reactivity in human branched-chain alpha-ketoacid dehydrogenase". Structure. 14 (2): 287–98. doi:10.1016/j.str.2005.10.009. PMID 16472748.
- Li J, Machius M, Chuang JL, et al. (2007). "The two active sites in human branched-chain alpha-keto acid dehydrogenase operate independently without an obligatory alternating-site mechanism". J. Biol. Chem. 282 (16): 11904–13. doi:10.1074/jbc.M610843200. PMID 17329260.
External links
- Human BCKDHB genome location and BCKDHB gene details page in the UCSC Genome Browser.
- v
- t
- e
-
 1dtw: HUMAN BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE
1dtw: HUMAN BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE -
 1ols: ROLES OF HIS291-ALPHA AND HIS146-BETA' IN THE REDUCTIVE ACYLATION REACTION CATALYZED BY HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE
1ols: ROLES OF HIS291-ALPHA AND HIS146-BETA' IN THE REDUCTIVE ACYLATION REACTION CATALYZED BY HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE -
 1olu: ROLES OF HIS291-ALPHA AND HIS146-BETA' IN THE REDUCTIVE ACYLATION REACTION CATALYZED BY HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE
1olu: ROLES OF HIS291-ALPHA AND HIS146-BETA' IN THE REDUCTIVE ACYLATION REACTION CATALYZED BY HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE -
 1olx: ROLES OF HIS291-ALPHA AND HIS146-BETA' IN THE REDUCTIVE ACYLATION REACTION CATALYZED BY HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE
1olx: ROLES OF HIS291-ALPHA AND HIS146-BETA' IN THE REDUCTIVE ACYLATION REACTION CATALYZED BY HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE -
 1u5b: Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase
1u5b: Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase -
 1v11: CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE
1v11: CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE -
 1v16: CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE
1v16: CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE -
 1v1m: CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE
1v1m: CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE -
 1v1r: CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE
1v1r: CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE -
 1wci: REACTIVITY MODULATION OF HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE BY AN INTERNAL MOLECULAR SWITCH
1wci: REACTIVITY MODULATION OF HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE BY AN INTERNAL MOLECULAR SWITCH -
 1x7w: Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase
1x7w: Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase -
 1x7x: Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase
1x7x: Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase -
 1x7y: Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase
1x7y: Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase -
 1x7z: Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase
1x7z: Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase -
 1x80: Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase
1x80: Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase -
 2beu: REACTIVITY MODULATION OF HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE BY AN INTERNAL MOLECULAR SWITCH
2beu: REACTIVITY MODULATION OF HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE BY AN INTERNAL MOLECULAR SWITCH -
 2bev: REACTIVITY MODULATION OF HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE BY AN INTERNAL MOLECULAR SWITCH
2bev: REACTIVITY MODULATION OF HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE BY AN INTERNAL MOLECULAR SWITCH -
 2bew: REACTIVITY MODULATION OF HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE BY AN INTERNAL MOLECULAR SWITCH
2bew: REACTIVITY MODULATION OF HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE BY AN INTERNAL MOLECULAR SWITCH -
 2bfb: REACTIVITY MODULATION OF HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE BY AN INTERNAL MOLECULAR SWITCH
2bfb: REACTIVITY MODULATION OF HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE BY AN INTERNAL MOLECULAR SWITCH -
 2bfc: REACTIVITY MODULATION OF HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE BY AN INTERNAL MOLECULAR SWITCH
2bfc: REACTIVITY MODULATION OF HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE BY AN INTERNAL MOLECULAR SWITCH -
 2bfd: REACTIVITY MODULATION OF HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE BY AN INTERNAL MOLECULAR SWITCH
2bfd: REACTIVITY MODULATION OF HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE BY AN INTERNAL MOLECULAR SWITCH -
 2bfe: REACTIVITY MODULATION OF HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE BY AN INTERNAL MOLECULAR SWITCH
2bfe: REACTIVITY MODULATION OF HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE BY AN INTERNAL MOLECULAR SWITCH -
 2bff: REACTIVITY MODULATION OF HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE BY AN INTERNAL MOLECULAR SWITCH
2bff: REACTIVITY MODULATION OF HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE BY AN INTERNAL MOLECULAR SWITCH -
 2j9f: HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE-DECARBOXYLASE E1B
2j9f: HUMAN BRANCHED-CHAIN ALPHA-KETOACID DEHYDROGENASE-DECARBOXYLASE E1B
 | This article on a gene on human chromosome 6 is a stub. You can help Wikipedia by expanding it. |
- v
- t
- e