Proto-oncogene tyrosine-protein kinase Src
| SRC | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | SRC, ASV, SRC1, c-p60-Src, SRC proto-oncogene, non-receptor tyrosine kinase, THC6 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 190090 MGI: 98397 HomoloGene: 21120 GeneCards: SRC | ||||||||||||||||||||||||||||||||||||||||||||||||||
| EC number | 2.7.10.2 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Proto-oncogene tyrosine-protein kinase Src, also known as proto-oncogene c-Src, or simply c-Src (cellular Src; pronounced "sarc", as it is short for sarcoma), is a non-receptor tyrosine kinase protein that in humans is encoded by the SRC gene. It belongs to a family of Src family kinases and is similar to the v-Src (viral Src) gene of Rous sarcoma virus. It includes an SH2 domain, an SH3 domain and a tyrosine kinase domain. Two transcript variants encoding the same protein have been found for this gene.[5]
c-Src phosphorylates specific tyrosine residues in other tyrosine kinases. It plays a role in the regulation of embryonic development and cell growth. An elevated level of activity of c-Src is suggested to be linked to cancer progression by promoting other signals.[6] Mutations in c-Src could be involved in the malignant progression of colon cancer. c-Src should not be confused with CSK (C-terminal Src kinase), an enzyme that phosphorylates c-Src at its C-terminus and provides negative regulation of Src's enzymatic activity.
c-Src was originally discovered by American scientists J. Michael Bishop and Harold E. Varmus, for which they were awarded the 1989 Nobel Prize in Physiology or Medicine.[7]
Discovery
In 1979, J. Michael Bishop and Harold E. Varmus discovered that normal chickens possess a gene that is structurally closely related to v-Src.[8] The normal cellular gene was called c-src (cellular-src).[9] This discovery changed the current thinking about cancer from a model wherein cancer is caused by a foreign substance (a viral gene) to one where a gene that is normally present in the cell can cause cancer. It is believed that at one point an ancestral virus mistakenly incorporated the c-Src gene of its cellular host. Eventually this normal gene mutated into an abnormally functioning oncogene within the Rous sarcoma virus. Once the oncogene is transfected back into a chicken, it can lead to cancer.
Structure
There are 9 members of the Src family kinases: c-Src, Yes, Fyn, Fgr, Yrk, Lyn, Blk, Hck, and Lck.[10] The expression of these Src family members are not the same throughout all tissues and cell types. Src, Fyn and Yes are expressed ubiquitously in all cell types while the others are generally found in hematopoietic cells.[11][12][13][14]
c-Src is made up of 6 functional regions: Src homology 4 domain (SH4 domain), unique region, SH3 domain, SH2 domain, catalytic domain and short regulatory tail.[15] When Src is inactive, the phosphorylated tyrosine group at the 527 position interacts with the SH2 domain which helps the SH3 domain interact with the flexible linker domain and thereby keeps the inactive unit tightly bound. The activation of c-Src causes the dephosphorylation of the tyrosine 527. This induces long-range allostery via protein domain dynamics, causing the structure to be destabilized, resulting in the opening up of the SH3, SH2 and kinase domains and the autophosphorylation of the residue tyrosine 416.[16][17][18]
The autophosphorylation of Y416 as well as phosphorylation of selected Src substrates is enhanced through dimerization of c-Src.[19] The dimerization of c-Src is mediated by the interaction of the myristoylated N-terminal region of one partner and the kinase domain of another partner.[19] Both the N-terminally attached myristic acid and the peptide sequences of the unique region are involved in the interaction.[19] Given the versatility inherent in this intrinsically disordered region, its multisite phosphorylations, and its divergence within the family, the unique domain likely functions as a central signaling hub overseeing much of the enzymatic activities and unique functions of Src family kinases.[19]
c-Src can be activated by many transmembrane proteins that include: adhesion receptors, receptor tyrosine kinases, G-protein coupled receptors and cytokine receptors. Most studies have looked at the receptor tyrosine kinases and examples of these are platelet derived growth factor receptor (PDGFR) pathway and epidermal growth factor receptor (EGFR).
Src contains at least three flexible protein domains, which, in conjunction with myristoylation, can mediate attachment to membranes and determine subcellular localization.[20]
Function
This proto-oncogene may play a role in the regulation of embryonic development and cell growth.
When src is activated, it promotes survival, angiogenesis, proliferation and invasion pathways. It also regulates angiogenic factors and vascular permeability after focal cerebral ischemia-reperfusion,[21][22] and regulates matrix metalloproteinase-9 activity after intracerebral hemorrhage.[23]
Role in cancer
The activation of the c-Src pathway has been observed in about 50% of tumors from colon, liver, lung, breast and the pancreas.[24] Since the activation of c-Src leads to the promotion of survival, angiogenesis, proliferation and invasion pathways, the aberrant growth of tumors in cancers is observed. A common mechanism is that there are genetic mutations that result in the increased activity or the overexpression of the c-Src leading to the constant activation of the c-Src.
Colon cancer
The activity of c-Src has been best characterized in colon cancer. Researchers have shown that Src expression is 5 to 8 fold higher in premalignant polyps than normal mucosa.[25][26][27] The elevated c-Src levels have also been shown to have a correlation with advanced stages of the tumor, size of tumor, and metastatic potential of tumors.[28][29]
Breast cancer
EGFR activates c-Src while EGF also increases the activity of c-Src. In addition, overexpression of c-Src increases the response of EGFR-mediated processes. So both EGFR and c-Src enhance the effects of one another. Elevated expression levels of c-Src were found in human breast cancer tissues compared to normal tissues.[30][31][32]
Overexpression of Human Epidermal Growth Factor Receptor 2 (HER2), also known as erbB2, is correlated with a worse prognosis for breast cancer.[33][34] Thus, c-Src plays a key role in the tumor progression of breast cancers.
Prostate cancer
Members of the Src family kinases Src, Lyn and Fgr are highly expressed in malignant prostate cells compared to normal prostate cells.[35] When the primary prostate cells are treated with KRX-123, which is an inhibitor of Lyn, the cells in vitro were reduced in proliferation, migration and invasive potential.[36] So the use of a tyrosine kinase inhibitor is a possible way of reducing the progression of prostate cancers.
As a drug target
A number of tyrosine kinase inhibitors that target c-Src tyrosine kinase (as well as related tyrosine kinases) have been developed for therapeutic use.[37] One notable example is dasatinib which has been approved for the treatment of chronic myeloid leukemia (CML) and Philadelphia chromosome-positive (PH+) acute lymphocytic leukemia (ALL).[38] Dasatinib is also in clinical trials for the use in non-Hodgkin’s lymphoma, metastatic breast cancer and prostate cancer. Other tyrosine kinase inhibitor drugs that are in clinical trials include bosutinib,[39] bafetinib, Saracatinib(AZD-0530), XLl-999, KX01 and XL228.[6] HSP90 inhibitor NVP-BEP800 has been described to affect stability of Src tyrosine kinase and growth of T-cell and B-cell acute lymphoblastic leukemias. [40]
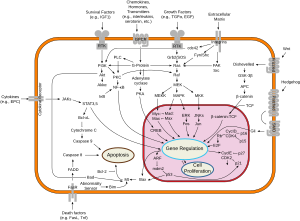
Interactions
Src (gene) has been shown to interact with the following signaling pathways:
Survival
Angiogenesis
Proliferation
Motility
Additional images
 |
|
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000197122 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000027646 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Entrez Gene: SRC v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)".
- ^ a b Wheeler DL, Iida M, Dunn EF (July 2009). "The role of Src in solid tumors". Oncologist. 14 (7): 667–78. doi:10.1634/theoncologist.2009-0009. PMC 3303596. PMID 19581523.
- ^ "The Nobel Prize in Physiology or Medicine 1989: J. Michael Bishop, Harold E. Varmus". Nobelprize.org. 1989-10-09.
for their discovery of 'the cellular origin of retroviral oncogenes'
- ^ Stehelin D, Fujita DJ, Padgett T, Varmus HE, Bishop JM (1977). "Detection and enumeration of transformation-defective strains of avian sarcoma virus with molecular hybridization". Virology. 76 (2): 675–84. doi:10.1016/0042-6822(77)90250-1. PMID 190771.
- ^ Oppermann H, Levinson AD, Varmus HE, Levintow L, Bishop JM (April 1979). "Uninfected vertebrate cells contain a protein that is closely related to the product of the avian sarcoma virus transforming gene (src)". Proc. Natl. Acad. Sci. U.S.A. 76 (4): 1804–8. Bibcode:1979PNAS...76.1804O. doi:10.1073/pnas.76.4.1804. PMC 383480. PMID 221907.
- ^ Thomas SM, Brugge JS (1997). "Cellular functions regulated by Src family kinases". Annu. Rev. Cell Dev. Biol. 13: 513–609. doi:10.1146/annurev.cellbio.13.1.513. PMID 9442882.
- ^ Cance WG, Craven RJ, Bergman M, Xu L, Alitalo K, Liu ET (December 1994). "Rak, a novel nuclear tyrosine kinase expressed in epithelial cells". Cell Growth Differ. 5 (12): 1347–55. PMID 7696183.
- ^ Lee J, Wang Z, Luoh SM, Wood WI, Scadden DT (January 1994). "Cloning of FRK, a novel human intracellular SRC-like tyrosine kinase-encoding gene". Gene. 138 (1–2): 247–51. doi:10.1016/0378-1119(94)90817-6. PMID 7510261.
- ^ Oberg-Welsh C, Welsh M (January 1995). "Cloning of BSK, a murine FRK homologue with a specific pattern of tissue distribution". Gene. 152 (2): 239–42. doi:10.1016/0378-1119(94)00718-8. PMID 7835707.
- ^ Thuveson M, Albrecht D, Zürcher G, Andres AC, Ziemiecki A (April 1995). "iyk, a novel intracellular protein tyrosine kinase differentially expressed in the mouse mammary gland and intestine". Biochem. Biophys. Res. Commun. 209 (2): 582–9. doi:10.1006/bbrc.1995.1540. PMID 7733928.
- ^ Arbesú M, Maffei M, Cordeiro TN, Teixeira JM, Pérez Y, Bernadó P, Roche S, Pons M (March 2017). "The Unique Domain Forms a Fuzzy Intramolecular Complex in Src Family Kinases". Structure. 25 (4): 630–640.e4. doi:10.1016/j.str.2017.02.011. PMID 28319009.
- ^ Cooper JA, Gould KL, Cartwright CA, Hunter T (March 1986). "Tyr527 is phosphorylated in pp60c-src: implications for regulation". Science. 231 (4744): 1431–4. Bibcode:1986Sci...231.1431C. doi:10.1126/science.2420005. PMID 2420005.
- ^ Okada M, Nakagawa H (December 1989). "A protein tyrosine kinase involved in regulation of pp60c-src function". J. Biol. Chem. 264 (35): 20886–93. doi:10.1016/S0021-9258(19)30019-5. PMID 2480346.
- ^ Nada S, Okada M, MacAuley A, Cooper JA, Nakagawa H (May 1991). "Cloning of a complementary DNA for a protein-tyrosine kinase that specifically phosphorylates a negative regulatory site of p60c-src". Nature. 351 (6321): 69–72. Bibcode:1991Natur.351...69N. doi:10.1038/351069a0. PMID 1709258. S2CID 4363527.
- ^ a b c d Spassov DS, Ruiz-Saenz A, Piple A, Moasser MM (Oct 2018). "A Dimerization Function in the Intrinsically Disordered N-Terminal Region of Src". Cell Rep. 25 (2): 6449–463. doi:10.1016/j.celrep.2018.09.035. PMC 6226010. PMID 30304684.
- ^ Kaplan JM, Varmus HE, Bishop JM (March 1990). "The src protein contains multiple domains for specific attachment to membranes". Molecular and Cellular Biology. 10 (3): 1000–9. doi:10.1128/mcb.10.3.1000. PMC 360952. PMID 1689455.
- ^ Zan L, Wu H, Jiang J, Zhao S, Song Y, Teng G, Li H, Jia Y, Zhou M, Zhang X, Qi J, Wang J (2011). "Temporal profile of Src, SSeCKS, and angiogenic factors after focal cerebral ischemia: correlations with angiogenesis and cerebral edema". Neurochem. Int. 58 (8): 872–9. doi:10.1016/j.neuint.2011.02.014. PMC 3100427. PMID 21334414.
- ^ Zan L, Zhang X, Xi Y, Wu H, Song Y, Teng G, Li H, Qi J, Wang J (2013). "Src regulates angiogenic factors and vascular permeability after focal cerebral ischemia-reperfusion". Neuroscience. 262 (3): 118–128. doi:10.1016/j.neuroscience.2013.12.060. PMC 3943922. PMID 24412374.
- ^ Zhao X, Wu T, Chang CF, et al. (2015). "Toxic role of prostaglandin E2 receptor EP1 after intracerebral hemorrhage in mice". Brain Behav. Immun. 46: 293–310. doi:10.1016/j.bbi.2015.02.011. PMC 4422065. PMID 25697396.
- ^ Dehm SM, Bonham K (April 2004). "SRC gene expression in human cancer: the role of transcriptional activation". Biochem. Cell Biol. 82 (2): 263–74. doi:10.1139/o03-077. PMID 15060621.
- ^ Bolen JB, Rosen N, Israel MA (November 1985). "Increased pp60c-src tyrosyl kinase activity in human neuroblastomas is associated with amino-terminal tyrosine phosphorylation of the src gene product". Proc. Natl. Acad. Sci. U.S.A. 82 (21): 7275–9. Bibcode:1985PNAS...82.7275B. doi:10.1073/pnas.82.21.7275. PMC 390832. PMID 2414774.
- ^ Cartwright CA, Kamps MP, Meisler AI, Pipas JM, Eckhart W (June 1989). "pp60c-src activation in human colon carcinoma". J. Clin. Invest. 83 (6): 2025–33. doi:10.1172/JCI114113. PMC 303927. PMID 2498394.
- ^ Talamonti MS, Roh MS, Curley SA, Gallick GE (January 1993). "Increase in activity and level of pp60c-src in progressive stages of human colorectal cancer". J. Clin. Invest. 91 (1): 53–60. doi:10.1172/JCI116200. PMC 329994. PMID 7678609.
- ^ Aligayer H, Boyd DD, Heiss MM, Abdalla EK, Curley SA, Gallick GE (January 2002). "Activation of Src kinase in primary colorectal carcinoma: an indicator of poor clinical prognosis". Cancer. 94 (2): 344–51. doi:10.1002/cncr.10221. PMID 11900220. S2CID 2103781.
- ^ Cartwright CA, Meisler AI, Eckhart W (January 1990). "Activation of the pp60c-src protein kinase is an early event in colonic carcinogenesis". Proc. Natl. Acad. Sci. U.S.A. 87 (2): 558–62. Bibcode:1990PNAS...87..558C. doi:10.1073/pnas.87.2.558. PMC 53304. PMID 2105487.
- ^ Ottenhoff-Kalff AE, Rijksen G, van Beurden EA, Hennipman A, Michels AA, Staal GE (September 1992). "Characterization of protein tyrosine kinases from human breast cancer: involvement of the c-src oncogene product". Cancer Res. 52 (17): 4773–8. PMID 1380891.
- ^ Biscardi JS, Belsches AP, Parsons SJ (April 1998). "Characterization of human epidermal growth factor receptor and c-Src interactions in human breast tumor cells". Mol. Carcinog. 21 (4): 261–72. doi:10.1002/(SICI)1098-2744(199804)21:4<261::AID-MC5>3.0.CO;2-N. PMID 9585256. S2CID 24236532.
- ^ Verbeek BS, Vroom TM, Adriaansen-Slot SS, Ottenhoff-Kalff AE, Geertzema JG, Hennipman A, Rijksen G (December 1996). "c-Src protein expression is increased in human breast cancer. An immunohistochemical and biochemical analysis". J. Pathol. 180 (4): 383–8. doi:10.1002/(SICI)1096-9896(199612)180:4<383::AID-PATH686>3.0.CO;2-N. PMID 9014858. S2CID 26892937.
- ^ Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL (January 1987). "Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene". Science. 235 (4785): 177–82. Bibcode:1987Sci...235..177S. doi:10.1126/science.3798106. PMID 3798106.
- ^ Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A (May 1989). "Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer". Science. 244 (4905): 707–12. Bibcode:1989Sci...244..707S. doi:10.1126/science.2470152. PMID 2470152.
- ^ Nam S, Kim D, Cheng JQ, Zhang S, Lee JH, Buettner R, Mirosevich J, Lee FY, Jove R (October 2005). "Action of the Src family kinase inhibitor, dasatinib (BMS-354825), on human prostate cancer cells". Cancer Res. 65 (20): 9185–9. doi:10.1158/0008-5472.CAN-05-1731. PMID 16230377.
- ^ Chang YM, Bai L, Yang I (2002). "Survey of Src activity and Src-related growth and migration in prostate cancer lines". Proc Am Assoc Cancer Res. 62: 2505a.
- ^ Musumeci F, Schenone S, Brullo C, Botta M (April 2012). "An update on dual Src/Abl inhibitors". Future Med Chem. 4 (6): 799–822. doi:10.4155/fmc.12.29. PMID 22530642.
- ^ Breccia M, Salaroli A, Molica M, Alimena G (2013). "Systematic review of dasatinib in chronic myeloid leukemia". OncoTargets Ther. 6: 257–65. doi:10.2147/OTT.S35360. PMC 3615898. PMID 23569389.
- ^ Amsberg GK, Koschmieder S (2013). "Profile of bosutinib and its clinical potential in the treatment of chronic myeloid leukemia". OncoTargets Ther. 6: 99–106. doi:10.2147/OTT.S19901. PMC 3594007. PMID 23493838.
- ^ Mshaik R, Simonet J, Georgievski A, Jamal L, Bechoua S, Ballerini P, Bellaye PS, Mlamla Z, Pais de Barros JP, Geissler A, Francin PJ, Girodon F, Garrido C, Quéré R (March 2021). "HSP90 inhibitor NVP-BEP800 affects stability of SRC kinases and growth of T-cell and B-cell acute lymphoblastic leukemias". Blood Cancer J. 3 (11): 61. doi:10.1038/s41408-021-00450-2. PMC 7973815. PMID 33737511.
External links
- src+Gene at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- src-Family+Kinases at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Proteopedia SRC - interactive 3D model of the structure of SRC
- Vega geneview
- Src Info with links in the Cell Migration Gateway Archived 2014-12-11 at the Wayback Machine
- Overview of all the structural information available in the PDB for UniProt: P12931 (Proto-oncogene tyrosine-protein kinase Src) at the PDBe-KB.
- v
- t
- e
-
 1a07: C-SRC (SH2 DOMAIN) COMPLEXED WITH ACE-MALONYL TYR-GLU-(N,N-DIPENTYL AMINE)
1a07: C-SRC (SH2 DOMAIN) COMPLEXED WITH ACE-MALONYL TYR-GLU-(N,N-DIPENTYL AMINE) -
 1a08: C-SRC (SH2 DOMAIN) COMPLEXED WITH ACE-DIFLUORO PHOSPHOTYR-GLU-(N,N-DIPENTYL AMINE)
1a08: C-SRC (SH2 DOMAIN) COMPLEXED WITH ACE-DIFLUORO PHOSPHOTYR-GLU-(N,N-DIPENTYL AMINE) -
 1a09: C-src (SH2 domain) complexed with ace-formyl phosphotyr-glu-(n,n-dipentyl amine)
1a09: C-src (SH2 domain) complexed with ace-formyl phosphotyr-glu-(n,n-dipentyl amine) -
 1a1a: C-SRC (SH2 DOMAIN WITH C188A MUTATION) COMPLEXED WITH ACE-FORMYL PHOSPHOTYR-GLU-(N,N-DIPENTYL AMINE)
1a1a: C-SRC (SH2 DOMAIN WITH C188A MUTATION) COMPLEXED WITH ACE-FORMYL PHOSPHOTYR-GLU-(N,N-DIPENTYL AMINE) -
 1a1b: C-SRC (SH2 DOMAIN) COMPLEXED WITH ACE-PHOSPHOTYR-GLU-(N,N-DIPENTYL AMINE)
1a1b: C-SRC (SH2 DOMAIN) COMPLEXED WITH ACE-PHOSPHOTYR-GLU-(N,N-DIPENTYL AMINE) -
 1a1c: C-SRC (SH2 DOMAIN) COMPLEXED WITH ACE-PHOSPHOTYR-GLU-(N-ME(-(CH2)3-CYCLOPENTYL))
1a1c: C-SRC (SH2 DOMAIN) COMPLEXED WITH ACE-PHOSPHOTYR-GLU-(N-ME(-(CH2)3-CYCLOPENTYL)) -
 1a1e: C-SRC (SH2 DOMAIN) COMPLEXED WITH ACE-PHOSPHOTYR-GLU-(3-BUTYLPIPERIDINE)
1a1e: C-SRC (SH2 DOMAIN) COMPLEXED WITH ACE-PHOSPHOTYR-GLU-(3-BUTYLPIPERIDINE) -
 1bkl: SELF-ASSOCIATED APO SRC SH2 DOMAIN
1bkl: SELF-ASSOCIATED APO SRC SH2 DOMAIN -
 1bkm: COCRYSTAL STRUCTURE OF D-AMINO ACID SUBSTITUTED PHOSPHOPEPTIDE COMPLEX
1bkm: COCRYSTAL STRUCTURE OF D-AMINO ACID SUBSTITUTED PHOSPHOPEPTIDE COMPLEX -
 1f1w: SRC SH2 THREF1TRP MUTANT COMPLEXED WITH THE PHOSPHOPEPTIDE S(PTR)VNVQN
1f1w: SRC SH2 THREF1TRP MUTANT COMPLEXED WITH THE PHOSPHOPEPTIDE S(PTR)VNVQN -
 1f2f: SRC SH2 THREF1TRP MUTANT
1f2f: SRC SH2 THREF1TRP MUTANT -
 1fmk: CRYSTAL STRUCTURE OF HUMAN TYROSINE-PROTEIN KINASE C-SRC
1fmk: CRYSTAL STRUCTURE OF HUMAN TYROSINE-PROTEIN KINASE C-SRC -
 1hcs: NMR STRUCTURE OF THE HUMAN SRC SH2 DOMAIN COMPLEX
1hcs: NMR STRUCTURE OF THE HUMAN SRC SH2 DOMAIN COMPLEX -
 1hct: NMR STRUCTURE OF THE HUMAN SRC SH2 DOMAIN COMPLEX
1hct: NMR STRUCTURE OF THE HUMAN SRC SH2 DOMAIN COMPLEX -
 1is0: Crystal Structure of a Complex of the Src SH2 Domain with Conformationally Constrained Peptide Inhibitor
1is0: Crystal Structure of a Complex of the Src SH2 Domain with Conformationally Constrained Peptide Inhibitor -
 1kc2: structure of the triple (Lys(beta)D3Ala, Asp(beta)C8Ala, AspCD2Ala) mutant of the Src SH2 domain bound to the PQpYEEIPI peptide
1kc2: structure of the triple (Lys(beta)D3Ala, Asp(beta)C8Ala, AspCD2Ala) mutant of the Src SH2 domain bound to the PQpYEEIPI peptide -
 1ksw: Structure of Human c-Src Tyrosine Kinase (Thr338Gly Mutant) in Complex with N6-benzyl ADP
1ksw: Structure of Human c-Src Tyrosine Kinase (Thr338Gly Mutant) in Complex with N6-benzyl ADP -
 1nlo: STRUCTURE OF SIGNAL TRANSDUCTION PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE
1nlo: STRUCTURE OF SIGNAL TRANSDUCTION PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE -
 1nlp: STRUCTURE OF SIGNAL TRANSDUCTION PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE
1nlp: STRUCTURE OF SIGNAL TRANSDUCTION PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE -
 1nzl: Crystal Structure of Src SH2 domain bound to doubly phosphorylated peptide PQpYEpYIPI
1nzl: Crystal Structure of Src SH2 domain bound to doubly phosphorylated peptide PQpYEpYIPI -
 1nzv: Crystal Structure of Src SH2 domain bound to doubly phosphorylated peptide PQpYIpYVPA
1nzv: Crystal Structure of Src SH2 domain bound to doubly phosphorylated peptide PQpYIpYVPA -
 1o41: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU78300.
1o41: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU78300. -
 1o42: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU81843.
1o42: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU81843. -
 1o43: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU82129.
1o43: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU82129. -
 1o44: Crystal structure of sh2 in complex with ru85052
1o44: Crystal structure of sh2 in complex with ru85052 -
 1o45: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU84687.
1o45: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU84687. -
 1o46: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU90395.
1o46: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU90395. -
 1o47: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU82209.
1o47: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU82209. -
 1o48: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU85053.
1o48: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU85053. -
 1o49: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU85493.
1o49: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU85493. -
 1o4a: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU82197.
1o4a: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU82197. -
 1o4b: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU83876.
1o4b: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU83876. -
 1o4c: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH PHOSPHATE.
1o4c: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH PHOSPHATE. -
 1o4d: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU78262.
1o4d: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU78262. -
 1o4e: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU78299.
1o4e: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU78299. -
 1o4f: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU79073.
1o4f: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU79073. -
 1o4g: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH DPI59.
1o4g: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH DPI59. -
 1o4h: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU79072.
1o4h: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU79072. -
 1o4i: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH PAS219.
1o4i: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH PAS219. -
 1o4j: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH ISO24.
1o4j: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH ISO24. -
 1o4k: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH PASBN.
1o4k: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH PASBN. -
 1o4l: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH FRAGMENT2.
1o4l: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH FRAGMENT2. -
 1o4m: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH MALONICACID.
1o4m: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH MALONICACID. -
 1o4n: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH OXALIC ACID.
1o4n: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH OXALIC ACID. -
 1o4o: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH PHENYLPHOSPHATE.
1o4o: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH PHENYLPHOSPHATE. -
 1o4p: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU78791.
1o4p: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU78791. -
 1o4q: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU79256.
1o4q: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU79256. -
 1o4r: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU78783.
1o4r: CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU78783. -
 1p13: Crystal Structure of the Src SH2 Domain Complexed with Peptide (SDpYANFK)
1p13: Crystal Structure of the Src SH2 Domain Complexed with Peptide (SDpYANFK) -
 1prl: TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS
1prl: TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS -
 1prm: TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS
1prm: TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS -
 1qwe: C-SRC SH3 DOMAIN COMPLEXED WITH LIGAND APP12
1qwe: C-SRC SH3 DOMAIN COMPLEXED WITH LIGAND APP12 -
 1qwf: C-SRC SH3 DOMAIN COMPLEXED WITH LIGAND VSL12
1qwf: C-SRC SH3 DOMAIN COMPLEXED WITH LIGAND VSL12 -
 1rlp: TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS
1rlp: TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS -
 1rlq: TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS
1rlq: TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS -
 1sha: CRYSTAL STRUCTURE OF THE PHOSPHOTYROSINE RECOGNITION DOMAIN SH2 OF V-SRC COMPLEXED WITH TYROSINE-PHOSPHORYLATED PEPTIDES
1sha: CRYSTAL STRUCTURE OF THE PHOSPHOTYROSINE RECOGNITION DOMAIN SH2 OF V-SRC COMPLEXED WITH TYROSINE-PHOSPHORYLATED PEPTIDES -
 1shb: CRYSTAL STRUCTURE OF THE PHOSPHOTYROSINE RECOGNITION DOMAIN SH2 OF V-SRC COMPLEXED WITH TYROSINE-PHOSPHORYLATED PEPTIDES
1shb: CRYSTAL STRUCTURE OF THE PHOSPHOTYROSINE RECOGNITION DOMAIN SH2 OF V-SRC COMPLEXED WITH TYROSINE-PHOSPHORYLATED PEPTIDES -
 1shd: PEPTIDE INHIBITORS OF SRC SH3-SH2-PHOSPHOPROTEIN INTERACTIONS
1shd: PEPTIDE INHIBITORS OF SRC SH3-SH2-PHOSPHOPROTEIN INTERACTIONS -
 1skj: COCRYSTAL STRUCTURE OF UREA-SUBSTITUTED PHOSPHOPEPTIDE COMPLEX
1skj: COCRYSTAL STRUCTURE OF UREA-SUBSTITUTED PHOSPHOPEPTIDE COMPLEX -
 1spr: BINDING OF A HIGH AFFINITY PHOSPHOTYROSYL PEPTIDE TO THE SRC SH2 DOMAIN: CRYSTAL STRUCTURES OF THE COMPLEXED AND PEPTIDE-FREE FORMS
1spr: BINDING OF A HIGH AFFINITY PHOSPHOTYROSYL PEPTIDE TO THE SRC SH2 DOMAIN: CRYSTAL STRUCTURES OF THE COMPLEXED AND PEPTIDE-FREE FORMS -
 1sps: BINDING OF A HIGH AFFINITY PHOSPHOTYROSYL PEPTIDE TO THE SRC SH2 DOMAIN: CRYSTAL STRUCTURES OF THE COMPLEXED AND PEPTIDE-FREE FORMS
1sps: BINDING OF A HIGH AFFINITY PHOSPHOTYROSYL PEPTIDE TO THE SRC SH2 DOMAIN: CRYSTAL STRUCTURES OF THE COMPLEXED AND PEPTIDE-FREE FORMS -
 1srl: 1H AND 15N ASSIGNMENTS AND SECONDARY STRUCTURE OF THE SRC SH3 DOMAIN
1srl: 1H AND 15N ASSIGNMENTS AND SECONDARY STRUCTURE OF THE SRC SH3 DOMAIN -
 1srm: 1H AND 15N ASSIGNMENTS AND SECONDARY STRUCTURE OF THE SRC SH3 DOMAIN
1srm: 1H AND 15N ASSIGNMENTS AND SECONDARY STRUCTURE OF THE SRC SH3 DOMAIN -
 1y57: Structure of unphosphorylated c-Src in complex with an inhibitor
1y57: Structure of unphosphorylated c-Src in complex with an inhibitor -
 1yi6: C-term tail segment of human tyrosine kinase (258-533)
1yi6: C-term tail segment of human tyrosine kinase (258-533) -
 1yoj: Crystal structure of Src kinase domain
1yoj: Crystal structure of Src kinase domain -
 1yol: Crystal structure of Src kinase domain in complex with CGP77675
1yol: Crystal structure of Src kinase domain in complex with CGP77675 -
 1yom: Crystal structure of Src kinase domain in complex with Purvalanol A
1yom: Crystal structure of Src kinase domain in complex with Purvalanol A -
 2bdf: Src kinase in complex with inhibitor AP23451
2bdf: Src kinase in complex with inhibitor AP23451 -
 2bdj: Src kinase in complex with inhibitor AP23464
2bdj: Src kinase in complex with inhibitor AP23464 -
 2h8h: Src kinase in complex with a quinazoline inhibitor
2h8h: Src kinase in complex with a quinazoline inhibitor -
 2hwo: Crystal structure of Src kinase domain in complex with covalent inhibitor
2hwo: Crystal structure of Src kinase domain in complex with covalent inhibitor -
 2hwp: Crystal structure of Src kinase domain in complex with covalent inhibitor PD168393
2hwp: Crystal structure of Src kinase domain in complex with covalent inhibitor PD168393 -
 2oiq: Crystal Structure of chicken c-Src kinase domain in complex with the cancer drug imatinib.
2oiq: Crystal Structure of chicken c-Src kinase domain in complex with the cancer drug imatinib. -
 2ptk: CHICKEN SRC TYROSINE KINASE
2ptk: CHICKEN SRC TYROSINE KINASE -
 2src: CRYSTAL STRUCTURE OF HUMAN TYROSINE-PROTEIN KINASE C-SRC, IN COMPLEX WITH AMP-PNP
2src: CRYSTAL STRUCTURE OF HUMAN TYROSINE-PROTEIN KINASE C-SRC, IN COMPLEX WITH AMP-PNP